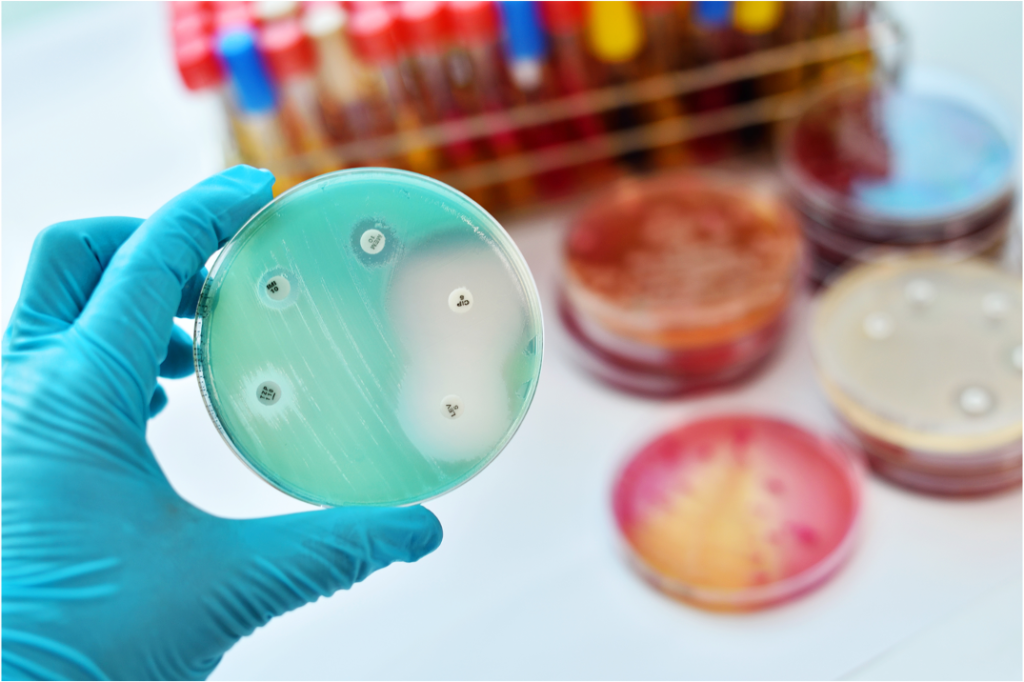
Natural products
Bacteria produce a wealth of small organic molecules to interact with their local environment. Many of these natural products have promising bioactivities and can be exploited in human and veterinary medicine for the treatment of a wide variety of ailments such as infectious diseases and cancer. Traditionally, natural products have been identified through extensive bioactivity-guided screening efforts focusing on few talented producer phyla that are frequently found in soil samples. These restrictions have resulted in the recurring discovery of already known natural products. As a result, the discovery pace of truly novel natural product scaffolds has slowed down significantly in the past decades. To circumvent the time-consuming rediscovery of known metabolites and identify truly novel natural products in the post-genomics era, we are working on the development of novel natural product discovery workflows and optimize existing strategies for their application in modern natural product discovery.

Genome Mining
Genes involved in the biosynthesis of a natural product are usually encoded in close vicinity – or clustered – in the genome of bacteria and core biosynthetic genes of a natural product class are usually characterized by a certain level of homology based on which they can be identified in the genome even though the product of the biosynthetic pathways might not have any apparent similarity. Despite the development of highly sophisticated tools, some natural product biosynthetic pathways remain unrecognized by state-of-the-art bioinformatic platforms. Others are recognized but encode proteins that are responsible for cryptic transformations.
Our goal is to identify novel families of natural product biosynthetic gene clusters in the ever-increasing wealth of publically available bacterial genomes and to characterize the associated products. Based on biosynthetic insights obtained from these studies, we develop bioinformatic tools for the identification of pathways that follow similar biosynthetic principles and predict the structures of the associated metabolites.

Chemical Ecology
Traditionally, natural products have been isolated by bioactivity-guided fractionation from extracts obtained from a few genera of talented secondary metabolite producers. This simple workflow has fueled the “Golden Age of Antibiotics” between 1950s and 1970s. Since the low hanging fruits have been picked, sticking to the same few talented producer phyla and sample types has resulted in the frequent rediscovery of known metabolites. We focus our efforts on poorly studied ecological niches that are shaped by natural products with a certain bioactivity. Our hypothesis-driven, function-first approach for the identification of bioactive natural products based on simple bioassays is an adaptation of the traditional bioactivity-guided fractionation workflow. In addition to the smart selection of an ecological niche to study and a robust bioassay these studies are complemented by imaging mass spectrometry to identify the metabolite responsible for the observed bioactivity prior to the isolation and genome sequencing as two means of early dereplication.
Natural Product Biosynthesis
Overlooked families and poorly studied classes of natural products frequently harbor genes that encode proteins that catalyze cryptic or unpredictable transformations. Studying these transformations can provide valuable insights into the biosynthesis of an entire family or class of natural products. These insights pave the way towards establishing universal biosynthetic principles for a natural product class or family and are a first step towards the design of novel, non-natural biosynthetic pathways.
Moreover, we use retrobiosynthetic analysis to identify the biosynthetic gene clusters for characterized natural products that are likely produced by non-canonical biosynthetic pathways harboring unusual biosynthetic enzymes or likely arising from spontaneous reactions.
Unusual proteins might be employed in chemoenzymatic syntheses to obtain difficult-to-synthesize chemical products and insights into the spontaneous formation of complex natural products might inspire biomimetic syntheses of these compounds.
