
Genome sequencing
As part of the LOEWE Center for Translational Biodiversity Genomics the Helfrich Lab uses state-of-the-art sequencing platforms (e.g. PacBio Sequel II) to sequence individual bacterial species, members of ecological niches and microbiomes of rare eukaryotes. Moreover, we assemble bacterial genomes from bacterial contaminants from eukaryotic genome sequencing projects and mine the assembled genomes for their natural product biosynthetic potential.

Bioinformatics
The Helfrich Lab develops genome mining platforms for the annotation of overlooked natural product families. The corresponding metabolites are characterized and the obtained biosynthetic insights used to deduce biosynthetic principles that are utilized for the development of algorithms for the structural prediction of the associated metabolites from genome sequence information.

Synthetic Biology
The Helfrich Lab employs and develops tools, and optimized host strains for the refactoring and heterologous expression of biosynthetic pathways, and the generation of small libraries of non-natural specialized metabolites. Our technology allow the characterization of biosynthetic pathways without the need of knocking out individual genes.

Medium-throughput Bioassays
The Helfrich Lab uses and develops high-throughput bioassays to characterize the potential of entire strain collections of bacterial isolates from specific ecological niches to produce natural products with antibacterial and antifungal properties. Simple methods are being developed to screen against multiple microbes simultaneously.
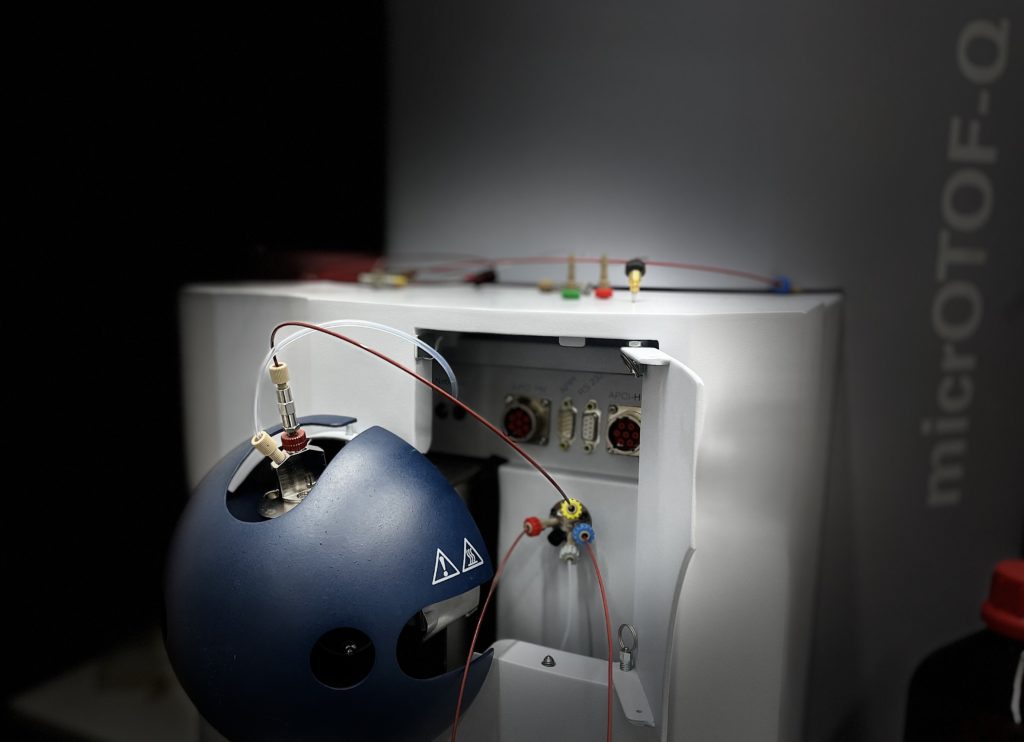
Metabolomics
The Helfrich Lab uses and develops chemoinformatics workflows for the analysis of bacterial secondary metabolites. We are equipped with analytical, semipreparative, and preparative HPLC setups connected to high and low resolution mass spectrometers. Complex extracts are fractionated and metabolites of interest purified by HPLC and their structures determined by NMR.
